MyGenostics Virus Genome Capture System
Product Overview
A virus is a small infectious agent that replicates only inside the living cells of other organisms. Infection of viruses could lead to various human diseases such as influenza, cold, AIDS and cancer. MyGenostics Virus Genome capture is designed to enrich genomes of different viruses as well as their subtypes from different types of samples of hosts, which could be further analyzed through Next-generation-sequencing platforms. MyGenostics virus genome capture system could effectively help researchers to identify different types of viruses, analyze mutations on the viruses and recognize the site of integration of virus genome into host’s genome, providing a new insight into medical research especially in cancer area.
System Workflow
.png)
Technical characteristics
Virus Genome Capture Series
.png)
MyGenostics also provide customerized virus genome capture service.
Sequencing Strategy
100PE, 125PE,150PE sequencing
Average sequencing depth: 500X
Data Analysis
Basic Analysis£ºHBV, HPV, EBV genome alignment, different types of genome, viruses genotyping, viruses integration analysis, virus mutation analysis.
Advanced Analysis: MyGenostics can provide customized analysis service such as Phylogenetic analysis, gene prediction.
Project time cycle
30 working days after verification of sample quality (without data analysis)
Additional 5~15 working days for data analysis
Case review
Overview:
This study employed MyGenostics EBV genome capture system to analyze the genomic diversity of EBV genomes isolated from primary gastric carcinoma biopsy.
Methodology:
206 gastric carcinoma specimens were assessed using in situ hybridization in order to identify EBV-associated gastric carcinoma sample. Then, the researcher employed Mygenostics EBV Genome Capture to extract EBV genome for sequencing analysis on 9 EBV-associated gastric carcinoma biopsy specimens.
Results:
It demonstrated that EBVaGC1 to 9 were most closely related to the GD1 strain. Phylogenetic analysis revealed the greatest divergence from the type 2 strain AG876. Compared with the reference EBV strain GD1, they harbored 961 variations in total, including 919 substitutions, 23 insertions, and 19 deletions. Moreover, 2 interstrain recombinants at the EBNA2 locus were identified. Some T-cell epitope sequences in EBNA1 and LMP2A genes showed extensive variation across strains.
Genetic Variations among GC-EBV strains
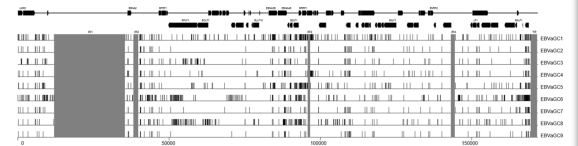
Pholygenetic trees of the whole EBV genomes and nucleotide sequences of EBNA1, EBNA2 and LMP1 genes
.png)
Conclusion:
This group reported the first genome-wide view of sequence variation of EBV isolated from primary EBVaGC biopsy specimens, which might serve as an effective method for further understanding the genomic variations contribute to EBVaGC carcinogenesis and treatment.
Reference
Liu Y, Yang W, Pan Y£¬Ji J, Lu Z, Ke Y (2015) Genome-wide analysis of Epstein-Barr virus(EBV) isolated from EBV-associated gastric carcinoma (EBVaGC). Oncotarget 2015.12.24